QuickStart
Kohleth Chia
2018-02-23
Preeliminaries
This is an R package so I assume you are already familiar with R.
Throughout this guide we will be using the dataset kinematics which comes with this package.
Quick Start
The functions in this package assume the kinematics data is already loaded in R, in the wide format. Specifically, the kinematics data.frame should contain at least columns curve_id, joint, plane, 0,1,2,...,100. Each row in the dataframe represent a kinematics curve on a particular joint and plane, identified by field curve_id, with data at 101 time points 0,1,2,...,100.
The package ships with an example data kinematics.
library(gaitFeature)
library(dplyr)
library(tidyr)
kinematics
#> # A tibble: 180 x 106
#> curve_id joint side plane ofs `0` `1` `2` `3` `4`
#> <int> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1 Pel L sag 49.0 28.2 28.1 28.1 28.2 28.2
#> 2 2 Pel R sag 52.0 27.8 27.9 28.0 28.1 28.2
#> 3 3 Hip L sag 49.0 52.4 52.5 52.6 52.8 52.9
#> 4 4 Hip R sag 52.0 54.1 55.3 56.6 57.9 59.1
#> 5 5 Knee L sag 49.0 17.2 19.2 21.6 24.1 26.7
#> 6 6 Knee R sag 52.0 10.8 13.8 17.4 21.4 25.4
#> 7 7 Ankle L sag 49.0 - 1.92 - 1.01 0.201 1.65 3.25
#> 8 8 Ankle R sag 52.0 - 7.27 - 7.13 - 6.39 - 5.09 - 3.36
#> 9 9 FootPro~ L sag 49.0 -95.2 -94.3 -93.3 -92.3 -91.5
#> 10 10 FootPro~ R sag 52.0 -99.3 -97.6 -96.0 -94.7 -93.6
#> # ... with 170 more rows, and 96 more variables: `5` <dbl>, `6` <dbl>,
#> # `7` <dbl>, `8` <dbl>, `9` <dbl>, `10` <dbl>, `11` <dbl>, `12` <dbl>,
#> # `13` <dbl>, `14` <dbl>, `15` <dbl>, `16` <dbl>, `17` <dbl>,
#> # `18` <dbl>, `19` <dbl>, `20` <dbl>, `21` <dbl>, `22` <dbl>,
#> # `23` <dbl>, `24` <dbl>, `25` <dbl>, `26` <dbl>, `27` <dbl>,
#> # `28` <dbl>, `29` <dbl>, `30` <dbl>, `31` <dbl>, `32` <dbl>,
#> # `33` <dbl>, `34` <dbl>, `35` <dbl>, `36` <dbl>, `37` <dbl>,
#> # `38` <dbl>, `39` <dbl>, `40` <dbl>, `41` <dbl>, `42` <dbl>,
#> # `43` <dbl>, `44` <dbl>, `45` <dbl>, `46` <dbl>, `47` <dbl>,
#> # `48` <dbl>, `49` <dbl>, `50` <dbl>, `51` <dbl>, `52` <dbl>,
#> # `53` <dbl>, `54` <dbl>, `55` <dbl>, `56` <dbl>, `57` <dbl>,
#> # `58` <dbl>, `59` <dbl>, `60` <dbl>, `61` <dbl>, `62` <dbl>,
#> # `63` <dbl>, `64` <dbl>, `65` <dbl>, `66` <dbl>, `67` <dbl>,
#> # `68` <dbl>, `69` <dbl>, `70` <dbl>, `71` <dbl>, `72` <dbl>,
#> # `73` <dbl>, `74` <dbl>, `75` <dbl>, `76` <dbl>, `77` <dbl>,
#> # `78` <dbl>, `79` <dbl>, `80` <dbl>, `81` <dbl>, `82` <dbl>,
#> # `83` <dbl>, `84` <dbl>, `85` <dbl>, `86` <dbl>, `87` <dbl>,
#> # `88` <dbl>, `89` <dbl>, `90` <dbl>, `91` <dbl>, `92` <dbl>,
#> # `93` <dbl>, `94` <dbl>, `95` <dbl>, `96` <dbl>, `97` <dbl>,
#> # `98` <dbl>, `99` <dbl>, `100` <dbl>The simplest use is to run the detectAll function which runs all the off-the-shelf feature detectors on the supplied dataframe.
detected=detectAll(kinematics)
detected%>%select(-(`0`:`100`))
#> # A tibble: 180 x 53
#> curve_id joint side plane ofs AIR DblB DFdec KFxatLdec HFxatICdec
#> <int> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1 Pel L sag 49.0 NA 1.00 NA NA NA
#> 2 2 Pel R sag 52.0 NA 1.00 NA NA NA
#> 3 3 Hip L sag 49.0 NA NA NA NA 0
#> 4 4 Hip R sag 52.0 NA NA NA NA 0
#> 5 5 Knee L sag 49.0 NA NA NA 0 NA
#> 6 6 Knee R sag 52.0 NA NA NA 0 NA
#> 7 7 Ankle L sag 49.0 NA NA 0 NA NA
#> 8 8 Ankle R sag 52.0 NA NA 0 NA NA
#> 9 9 Foot~ L sag 49.0 NA NA NA NA NA
#> 10 10 Foot~ R sag 52.0 NA NA NA NA NA
#> # ... with 170 more rows, and 43 more variables: Tiltdec <dbl>,
#> # TiltdecAndROMinc <dbl>, PkKFxdec <dbl>, DlyPkKFxdec <dbl>,
#> # DlyPkKFxat <dbl>, DlyPkKFx <dbl>, Desc2R <dbl>, DFatSwg <dbl>,
#> # EPS <dbl>, Abd <dbl>, AbdatSwn <dbl>, Add <dbl>, FtD <dbl>,
#> # KFxatMSinc <dbl>, H2R <dbl>, AddatStn <dbl>, HER <dbl>, HExtdec <dbl>,
#> # HypHFx <dbl>, HIR <dbl>, HypE <dbl>, DFinc <dbl>, KFxatICinc <dbl>,
#> # KFxatICincAndEE <dbl>, HExtatMSinc <dbl>, HFxinc <dbl>,
#> # HFxincAndROMdec <dbl>, HIRatLSinc <dbl>, MaxDFinc <dbl>,
#> # PROMinc <dbl>, PRotROMinc <dbl>, Tiltinc <dbl>,
#> # TiltincAndROMinc <dbl>, PFinc <dbl>, PkKFxat <dbl>, In <dbl>,
#> # iPP <dbl>, N1R <dbl>, Out <dbl>, ElevOrDep <dbl>, ProOrRe <dbl>,
#> # PrevROM <dbl>, S2R <dbl>detectAll returns the same data.frame as what is supplied, with extra columns – one for each feature. If the feature column is NA, it means that feature is not applicable to that curve (e.g. Ankle Internal Rotation (AIR) not relevant for the Pelvis curve.) If the field is 1 then that feature is detected, and 0 indicates the contrary.
The list of feature detectors that are called by detectAll is specified through the fDict argument. This argument should be a data.frame with column fn which list the names of the detectors to be called. Furthermore, it can have columns joint and plane which indicates which joint and plane a particular detector is related to. If fDict=NULL (default), it is generated on the spot by the makeFDict function.
makeFDict()
#> # A tibble: 48 x 5
#> fn joint plane fname featurename
#> <chr> <chr> <chr> <chr> <chr>
#> 1 .dtAIR Ankle tra AIR Ankle Internal Rotati~
#> 2 .dtDblBump Pel sag DblB Increased ROM (double~
#> 3 .dtDecDF Ankle sag DFdec Reduced Dorsiflexion
#> 4 .dtDecFxLoading Knee sag KFxatLdec Reduced flexion @ loa~
#> 5 .dtDecHipFxIC Hip sag HFxatICdec Decreased hip flexion~
#> 6 .dtDecPelTilt Pel sag Tiltdec Decreased pelvic tilt
#> 7 .dtDecPelTiltIncROM Pel sag TiltdecAndROMinc "Decreased Pelvic Til~
#> 8 .dtDecPkKnFx Knee sag PkKFxdec Decreased Peak Knee F~
#> 9 .dtDelayDecPkKnFx Knee sag DlyPkKFxdec Delayed + Decreased P~
#> 10 .dtDelayIncPkKnFx Knee sag DlyPkKFxat Delayed + Increased P~
#> # ... with 38 more rowsIf you are only interested at features that are a particular joint or plane, you can specify that in the detectAll call. But make sure the joint and plane value appears in the fDict data.frame.
detectAll(kinematics, joint="Hip", plane="sag")%>%
select(-(`0`:`100`))
#> # A tibble: 180 x 11
#> curve_id joint side plane ofs HFxatICdec HExtdec HypHFx HExtatMSinc
#> <int> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1 Pel L sag 49.0 NA NA NA NA
#> 2 2 Pel R sag 52.0 NA NA NA NA
#> 3 3 Hip L sag 49.0 0 0 0 0
#> 4 4 Hip R sag 52.0 0 0 0 0
#> 5 5 Knee L sag 49.0 NA NA NA NA
#> 6 6 Knee R sag 52.0 NA NA NA NA
#> 7 7 Ankle L sag 49.0 NA NA NA NA
#> 8 8 Ankle R sag 52.0 NA NA NA NA
#> 9 9 FootP~ L sag 49.0 NA NA NA NA
#> 10 10 FootP~ R sag 52.0 NA NA NA NA
#> # ... with 170 more rows, and 2 more variables: HFxinc <dbl>,
#> # HFxincAndROMdec <dbl>Note that all of kinematics will still be returned. So the above result is different to
kinematics%>%
filter(joint=="Hip",plane=="sag")%>%
detectAll(joint="Hip",plane="sag")%>%
select(-(`0`:`100`))
#> # A tibble: 12 x 11
#> curve_id joint side plane ofs HFxatICdec HExtdec HypHFx HExtatMSinc
#> <int> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 3 Hip L sag 49.0 0 0 0 0
#> 2 4 Hip R sag 52.0 0 0 0 0
#> 3 33 Hip L sag 48.0 0 0 0 0
#> 4 34 Hip R sag 54.0 0 0 0 0
#> 5 63 Hip L sag 50.0 0 0 0 0
#> 6 64 Hip R sag 49.0 0 0 0 0
#> 7 93 Hip L sag 50.0 0 0 0 0
#> 8 94 Hip R sag 50.0 0 0 0 0
#> 9 123 Hip L sag 50.0 0 0 0 0
#> 10 124 Hip R sag 51.0 0 0 0 0
#> 11 153 Hip L sag 50.0 0 0 0 0
#> 12 154 Hip R sag 49.0 0 0 0 0
#> # ... with 2 more variables: HFxinc <dbl>, HFxincAndROMdec <dbl>A (more) detailed explanation of why each feature is detected or not can be returned by setting detectAll(...,comp=TRUE) and inspecting the attributes.
z=detectAll(kinematics,joint="Hip",plane="sag",comp=TRUE)
head(attr(z,"comp"),1)
#> [[1]]
#> # A tibble: 12 x 3
#> curve_id `Cl:c(0)<2sd` HFxatICdec
#> <int> <lgl> <dbl>
#> 1 3 F 0
#> 2 4 F 0
#> 3 33 F 0
#> 4 34 F 0
#> 5 63 F 0
#> 6 64 F 0
#> 7 93 F 0
#> 8 94 F 0
#> 9 123 F 0
#> 10 124 F 0
#> 11 153 F 0
#> 12 154 F 0attr(.,"comp") returns one dataframe for each feature. The columns that begin with Cl: indicates it is a clause (component) of the feature. All clauses must be TRUE for a feature to be detected.
You can also call the individual detector directly.
.dtDecHipFxIC(kinematics)
#> # A tibble: 12 x 3
#> curve_id `Cl:c(0)<2sd` HFxatICdec
#> <int> <lgl> <dbl>
#> 1 3 F 0
#> 2 4 F 0
#> 3 33 F 0
#> 4 34 F 0
#> 5 63 F 0
#> 6 64 F 0
#> 7 93 F 0
#> 8 94 F 0
#> 9 123 F 0
#> 10 124 F 0
#> 11 153 F 0
#> 12 154 F 0Resampling
All functions in this package assume the kinematics curves are sampled at 101 time points. If this is not the case, there is a sampling function ?resamp101 which fits a periodic cubic spline to your existing curves, and then resample the 101 times points from it.
## x is a coarser version of kinematics
x=kinematics%>%
select(curve_id,`0`,`10`,`20`,`30`,`40`,`50`,`60`,`70`,`80`,`90`,`100`)
y=resamp101(x,`0`:`100`,plot=T)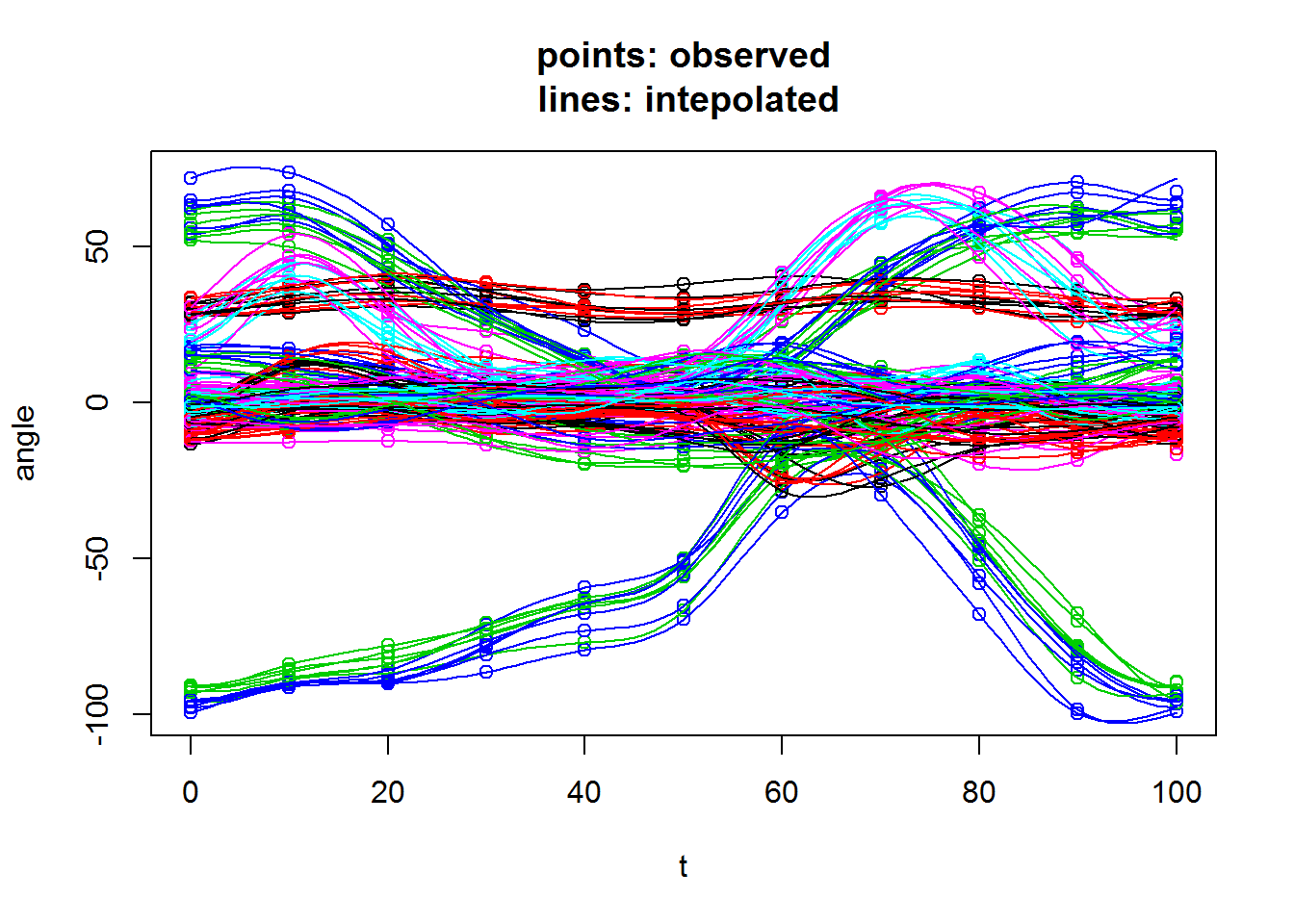
The plot=T argument plots the interpolated lines together with the observed points. It is recommended that you leave it as FALSE (default) if you are resampling many curves, because the plot will just be cluttered.